














If you work in cancer biomarker and target research, chances are you use data from The Cancer Genome Atlas (TCGA) to help you make discoveries. This comprehensive and coordinated effort helps accelerate our understanding of the molecular causes of cancer through genomic analyses, including large-scale genome sequencing. TCGA covers 33 types of cancer with multi-omics data, such as RNA-seq, DNA-seq, copy number, microRNA-seq and others. Detailed analyses of individual TCGA datasets, as well as pan-cancer meta-analysis, have revealed new cancer subtypes with important therapeutic implications. A key value here is the TCGA metadata. TCGA samples include extensive clinical metadata for diverse cancers. However, inconsistent terminology and formatting limit the utility of these data for pan-cancer analyses.
TCGA data within QIAGEN OmicSoft OncoLand is rigorously curated by experts who apply extensive ontologies and formatting rules to maximize consistency. This allows researchers to more easily find and understand patient characteristics, discover related covariates and explore patterns of clinical parameters across cancers in the context of multi-omics data. QIAGEN OmicSoft established strict standards through our curation of over 600,000 disease-relevant ‘omics samples. QIAGEN OmicSoft Lands provide access to uniformly processed datasets, in-depth metadata curation and data exploration tools that enable quick insights from thousands of deeply-curated ‘omics studies across therapeutic areas. QIAGEN OmicSoft Lands centralize data from Gene Expression Omnibus (GEO), NCBI Sequence Read Archive (SRA), ArrayExpress, TCGA, Cancer Cell Line Encyclopedia (CCLE), Genotype-Tissue Expression (GTEx), Blueprint, International Cancer Genome Consortium (ICGC), Therapeutically Applicable Research to Generate Effective Treatments (TARGET) and others.
OmicSoft’s curation process for TCGA
To give you an idea of the extensive time and care QIAGEN curators invest in manual curation of public ‘omics data, they recently spent over 1400 hours performing a comprehensive update of TCGA metadata within QIAGEN OmicSoft OncoLand, reviewing over 1200 source files. Clinical metadata are now comprehensively documented to clarify the meaning of fields in alignment with the latest OmicSoft curation standards. When TCGA metadata fields are redundant or unclear, new field names are used to clarify meaning. In addition, new metadata from recent TCGA publications are matched to TCGA data to apply recent discoveries about molecularly defined cancer subtypes.
At the core of the OmicSoft curation process, curators apply scientific expertise, controlled vocabulary and standardized formatting to all applicable metadata, either as a Fully Controlled Field (key clinical parameters use terms from QIAGEN-defined ontologies) or a Format Controlled Field (where a QIAGEN OmicSoft ontology is not applicable, terms are formatted consistently to maximize uniformity from semi-structured data). This means you can quickly and easily find all applicable samples using simplified search criteria.
Unification of related TCGA metadata fields
With data submissions from dozens of labs, groups adopt inconsistent standards to represent the same data. Where possible, OmicSoft curators identified hundreds of columns containing the same information for various tumors and combined the data into unified columns to enhance pan-cancer analyses and computational analysis. As an example, the cancer diagnosis of a first-degree family member with a history of cancer was captured in TCGA across five fields from four cancers; QIAGEN OmicSoft TCGA curation unites these into the single field “Family History [Cancer] [Type]”.
Synonymous terms and typographical errors
QIAGEN OmicSoft manually applies extensive treatment ontologies to ensure proper and unambiguous labeling of samples with treatment terms. Because of the many submitting groups, different standards were used for well-established terms, such as drug and radiation treatments, with occasional typos escaping submitter quality control checks. For example, over 20 different terms were used to describe treatment with doxorubicin!
Want to learn more about how you can boost your TCGA exploration to get quicker and more meaningful insights?
Read our white paper to get the full details of how QIAGEN OmicSoft OncoLand helps boost TCGA exploration. Download our unique and comprehensive metadata dictionary of clinical covariates to quickly discover the meaning of over 1000 relevant fields for deeper TCGA data exploration across cancers.
Learn more about the costs of free data in our industry report. Check out our infographic that details the various QIAGEN OmicSoft software tools for integrated ‘omics data, to see which solutions should help you transform your biomarker and target discovery.
New content, new projects, External Scripts improvements and more!
Recently-added Lands in OncoLand
OncoLand has added several new Lands over recent releases, be sure to check them out! If you do not see these Lands in your OncoLand collection, please contact your QIAGEN OmicSoft Server administrator to add the Lands to your server.
Body Maps
GTEx_B37 has 8711 new RNA-seq samples, and 16,964 total RNA-seq samples. GTEx_B38 is scheduled to be updated to GTEx V8 with 2020R2.
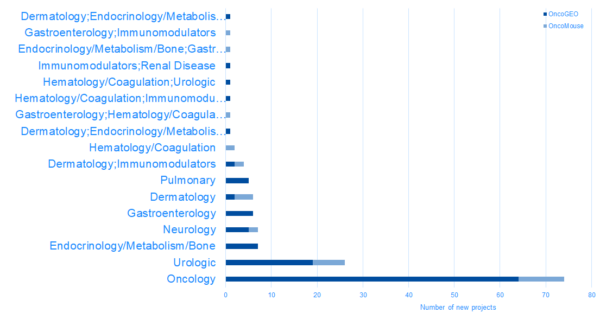
New projects in OncoLand 2020R1
New projects in DiseaseLand 2020R1
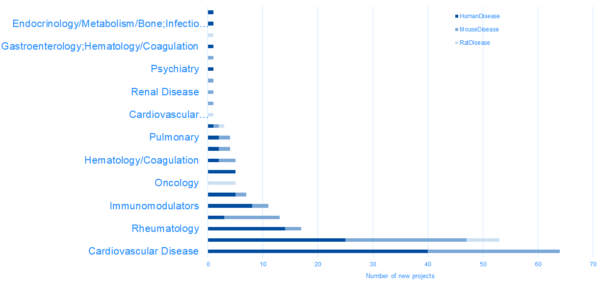
HumanDisease project highlights:
RatDisease project highlights:
MouseHumanDisease project highlights:
Array Suite is now QIAGEN OmicSoft Suite
This is purely a name change to reflect the wide variety of Omics data supported by QIAGEN OmicSoft.
OmicSoft Studio=Array Studio. OmicSoft Server=Array Server. OmicSoft Viewer=Array Viewer.<

Improved External Scripts with support for Docker technology and cloud analysis
OmicSoft Suite now supports External Scripts on AWS Cloud and can run analyses on Docker images. See some examples here.
These new capabilities substantially expand the options for OmicSoft Suite as an ‘omics data and analysis hub, allowing advanced users who would like to run third-party bioinformatics tools to do so from OmicSoft Suite, and even build pipelines to analyze data and import into OmicSoft projects. Talk with your account manager to learn more about some of the possibilities.
Updates to other OmicSoft Suite functions
Now you can download FASTQ files up to 100GB from the Short Read Archive. Previously, downloading FASTQ data from the NCBI Short Read Archive (SRA) database supported files only up to 20 GB.
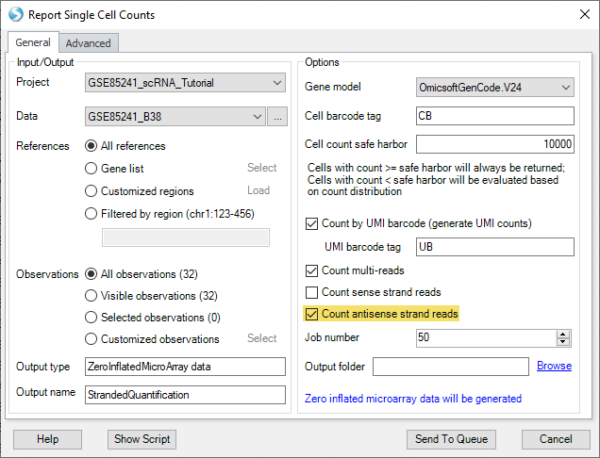
The “Single Cell Quantification” function now supports antisense strand reads (commonly generated from 5′ chemistry) in addition to sense strand reads (3′ chemistry), increasing the flexibility to support new workflows.
In “Map RNA-seq reads to genome“, there is a new option to pair input files in the order they were submitted, instead of pre-sorting the files by name. This is useful in exceptional situations, such as when data for the same sample are stored across multiple files in multiple directories, and the file names are identical between directories.
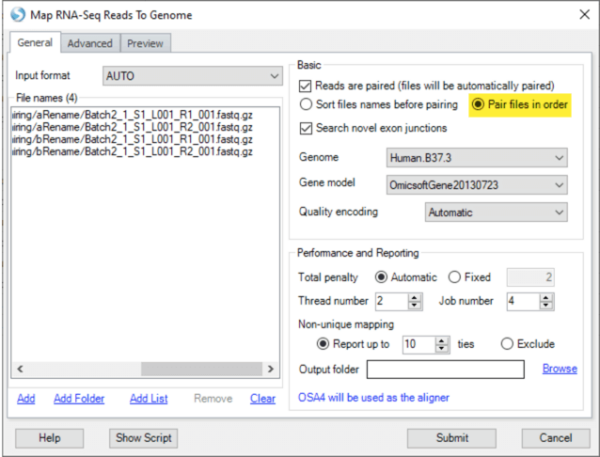
For example, if your data were run across multiple lanes, and the output files for Read1 are saved as “Batch2_1_S1_L001_R1_001.fastq.gz” in multiple directories (each directory holding data from one lane), you can ensure proper file pairing by specifying the order with “Add List” or during sample registration, and by selecting “pair files in order” when specifying alignment options.

Our recent virtual QIAGEN IPA/OmicSoft User Group Meetings that took place in April and May were both a great success. We hope you were able to join us! Review the recordings here.
Stay tuned for more QIAGEN IPA/OmicSoft User Group Meetings taking place later this year.
Learn more about the QIAGEN OmicSoft portfolio here.
OncoLand is a sophisticated oncology-focused database designed to accelerate cancer research. Integrating published research and large consortium cancer datasets, robust data visualization and discovery tools, OncoLand saves you valuable time and resources in your pursuit for actionable discoveries.
Start your free trial of OncoLand today!

To the many attendees and speakers: we can’t thank you enough for spending time with us in Cambridge, Mass.! For those who could not join us in person, below is a short summary about how OmicSoft’s content and software products are being used by pharma and biotech scientists to develop the next round of drug candidates and therapies. And in case you missed it, don’t forget to check out our day 1 highlights as well.
We were delighted to have Paul Jung, senior biology data scientist from AbbVie, give a presentation about how his group has utilized OmicSoft technology to build an ’omics data hub for his company. He laid out the challenge facing many IT teams in pharma and biotech: maximizing the value of disparate data sets, gathered with different platforms, and represented in diverse formats. His group is focused on enabling cross-platform analysis for microarray, proteomics, NGS, and chemical biology data. To start with, they addressed the need for centralized, secure storage; a controlled, common vocabulary; and flexible access control. They chose Array Studio in the cloud, developed a controlled vocabulary, and made it possible for AbbVie scientists to use custom annotations and to import external analysis files. Jung demonstrated some impressive applications of Array Studio, such as plotting compound potency versus ’omics data to power early discovery efforts. To date, he said this approach has been used to process more than 26,000 samples from AbbVie projects, and that his team has helped establish 14 internal Lands and seven internal clinical Lands data sets along the way.

QIAGEN’s Matt Newman offered a look at the custom curation services available for OmicSoft users. We have found that many customers would like additional content added to Lands for greater coverage of a certain disease or subject area, but they simply don’t have the internal resources to perform the strict and methodical curation necessary. It’s a tedious process — we can attest to that, since we do it every day! To help users get past this phase and into the more interesting science it facilitates, we can take on custom curation projects either for specific data sets or for a particular subject area. We’ll select and retrieve the most useful and relevant data, perform statistical modeling, extract metadata, establish fields, assign it to curators, edit the results, and then handle data merging and QC before publishing it to Lands. This way, users can have confidence that every bit of data they encounter in OmicSoft tools has been through the same rigorous curation, quality control, and review process for the best scientific results. Newman also noted that in 2018 we’ll be expanding these services to include analysis as well; this request has been frequently heard from customers at companies without lots of bioinformatics expertise.
Also, our own Nirav Amin presented several step-by-step case studies showing how to use OncoLand. Those can be viewed on our wiki.
We’ll be releasing videos and other materials to provide more details about the OmicSoft User Group Meeting. Stay tuned!