














New content, new projects, External Scripts improvements and more!
Recently-added Lands in OncoLand
OncoLand has added several new Lands over recent releases, be sure to check them out! If you do not see these Lands in your OncoLand collection, please contact your QIAGEN OmicSoft Server administrator to add the Lands to your server.
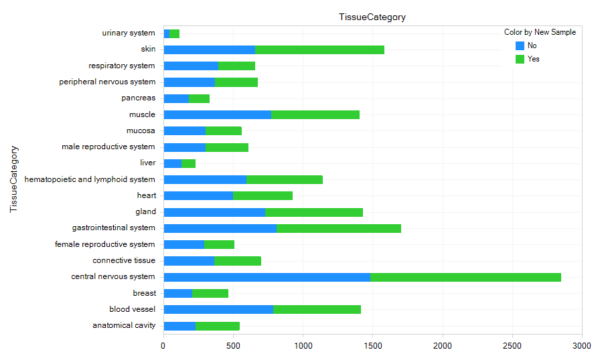
Body Maps
GTEx_B37 has 8711 new RNA-seq samples, and 16,964 total RNA-seq samples. GTEx_B38 is scheduled to be updated to GTEx V8 with 2020R2.
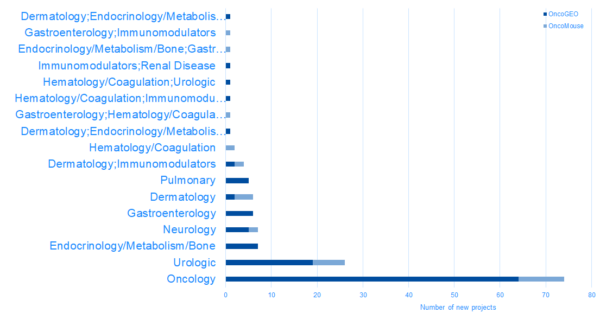
New projects in OncoLand 2020R1
New projects in DiseaseLand 2020R1
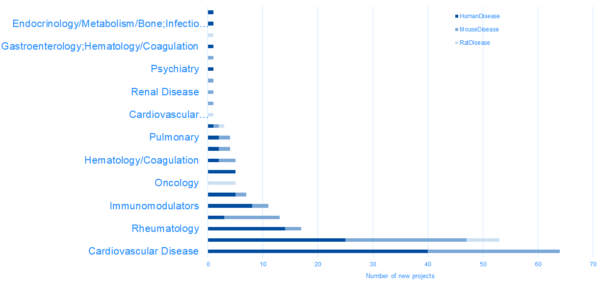
HumanDisease project highlights:
RatDisease project highlights:
MouseHumanDisease project highlights:
Array Suite is now QIAGEN OmicSoft Suite
This is purely a name change to reflect the wide variety of Omics data supported by QIAGEN OmicSoft.
OmicSoft Studio=Array Studio. OmicSoft Server=Array Server. OmicSoft Viewer=Array Viewer.<

Improved External Scripts with support for Docker technology and cloud analysis
OmicSoft Suite now supports External Scripts on AWS Cloud and can run analyses on Docker images. See some examples here.
These new capabilities substantially expand the options for OmicSoft Suite as an ‘omics data and analysis hub, allowing advanced users who would like to run third-party bioinformatics tools to do so from OmicSoft Suite, and even build pipelines to analyze data and import into OmicSoft projects. Talk with your account manager to learn more about some of the possibilities.
Updates to other OmicSoft Suite functions
Now you can download FASTQ files up to 100GB from the Short Read Archive. Previously, downloading FASTQ data from the NCBI Short Read Archive (SRA) database supported files only up to 20 GB.
The “Single Cell Quantification” function now supports antisense strand reads (commonly generated from 5′ chemistry) in addition to sense strand reads (3′ chemistry), increasing the flexibility to support new workflows.
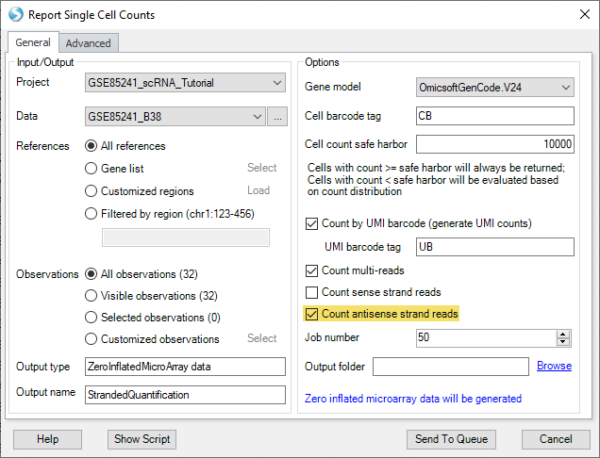
In “Map RNA-seq reads to genome“, there is a new option to pair input files in the order they were submitted, instead of pre-sorting the files by name. This is useful in exceptional situations, such as when data for the same sample are stored across multiple files in multiple directories, and the file names are identical between directories.
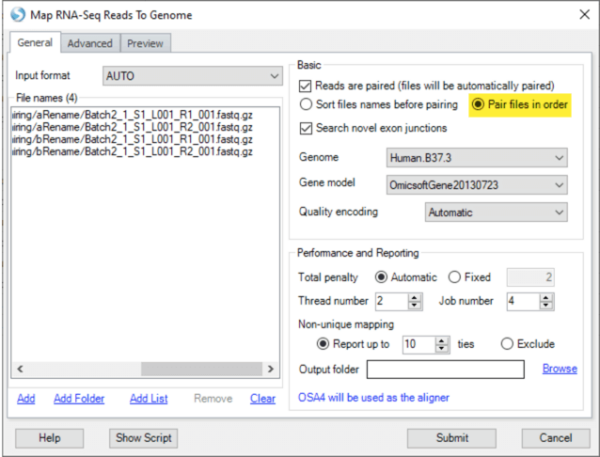
For example, if your data were run across multiple lanes, and the output files for Read1 are saved as “Batch2_1_S1_L001_R1_001.fastq.gz” in multiple directories (each directory holding data from one lane), you can ensure proper file pairing by specifying the order with “Add List” or during sample registration, and by selecting “pair files in order” when specifying alignment options.

Our recent virtual QIAGEN IPA/OmicSoft User Group Meetings that took place in April and May were both a great success. We hope you were able to join us! Review the recordings here.
Stay tuned for more QIAGEN IPA/OmicSoft User Group Meetings taking place later this year.
Learn more about the QIAGEN OmicSoft portfolio here.
OmicSoft, now a QIAGEN company, is excited to announce Array Suite 10.0, the ten-year anniversary release of its flagship software product. Array Suite provides the backbone of OmicSoft's software and data service offerings, including OncoLand, DiseaseLand and GeneticsLand. In the past ten years, Array Suite has helped numerous users from major pharma and biotech companies (as well as research institutions) accelerate their bioinformatics and genomics research.
See Jack Liu from OmicSoft present the top ten new features and more.
Founded in 2007, OmicSoft had a vision to focus on biomarker data management, visualization, and analysis. Array Suite (Array Studio and Array Server) differs from standard desktop solutions or open source solutions, with Array Studio providing the graphical user interface for NGS and OMIC analysis and visualization and Array Server providing the enterprise back-end solution for pipelines, project management, sample/file management, data storage and OMIC data warehouse (Land database). In January 2017, QIAGEN enhanced its portfolio with the acquisition of OmicSoft, allowing us to imagine new possibilities for integration with the larger QIAGEN bioinformatics portfolio. We will update everyone on these enhancements, and how they will benefit our users, in the near future.
"Although much has changed in the past ten years, in both software and the company itself, I'm proud that OmicSoft Corporation has remained unchanged in it's fundamental desire to implement useful tools driven by our customer's needs in the -OMICS space. I am confident that this will continue into the future with our acquisition by QIAGEN, and I look forward to many more years of Array Studio helping to drive exciting breakthroughs and research by our customers"
Matt Newman, VP Business Development
OmicSoft is extremely proud of it's customer-centric product development and customer support, and we look to continue this into the future, as we have for the past 10 years. With our latest update, this trend continues. Array Suite 10.0 includes revolutionary updates, with multiple technology breakthroughs including: Cloud-Based Lands, Single Cell RNA-Seq support, ENCODE integration and many other updates to both analytics and framework.
Here is a list of some of our exciting updates: