














Across oncology applications, from research to molecular testing and pharmaceutical development, the ability to identify potentially actionable genetic alterations and exploit the molecular vulnerabilities of cancer is becoming increasingly difficult. Due to the sporadic nature of somatic cancers, the number of detected variants is exponentially growing, challenging labs to confidently identify meaningful mutations that could influence or improve decisions at the point-of-care.
The Human Somatic Mutation Database (HSMD) is a new somatic database developed by QIAGEN that contains extensive genomic content relevant to solid tumors and hematological malignancies.
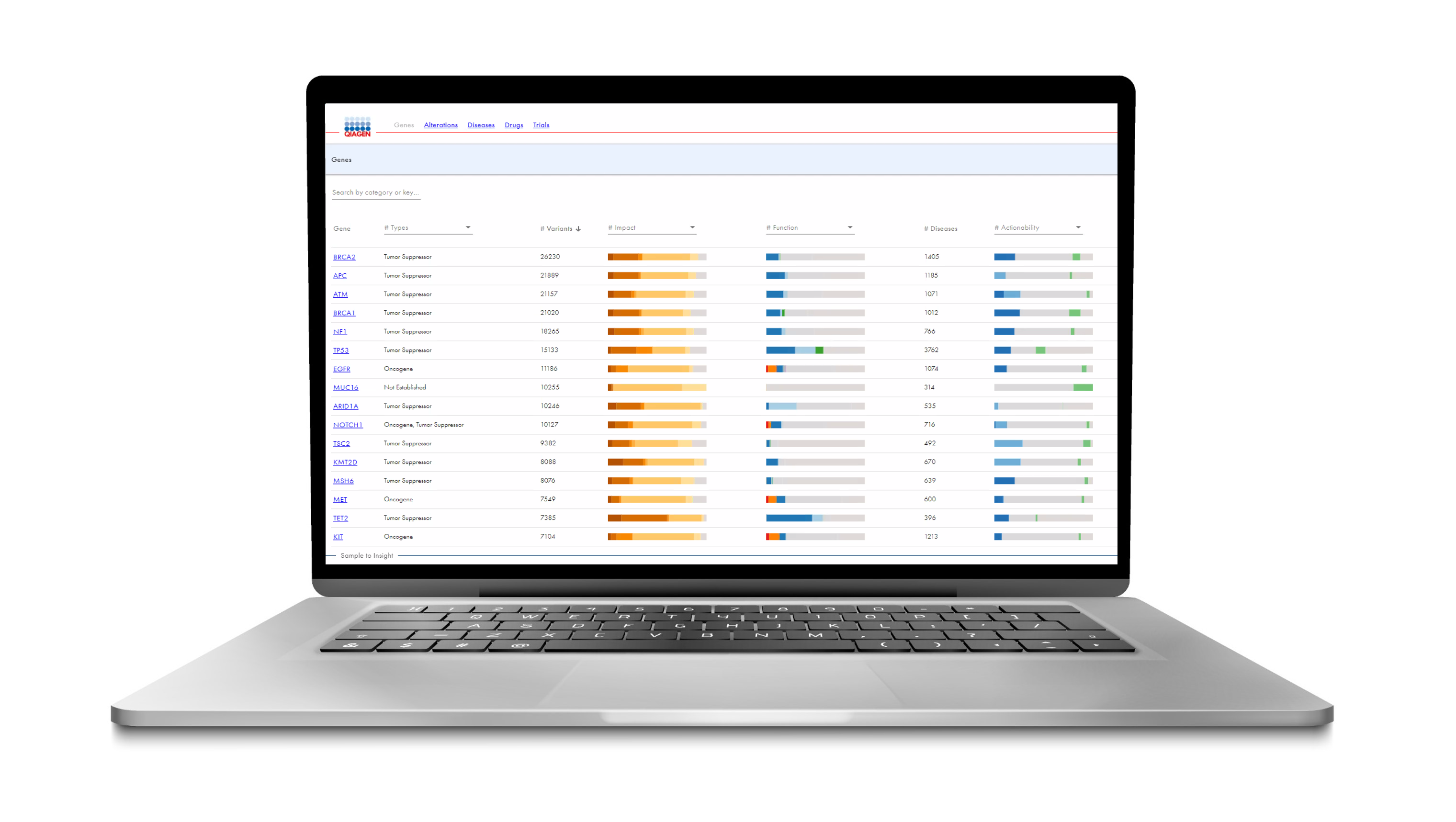
In the latest version of HSMD, the resource focuses on providing deep insight into small variants, such as SNVs, indels, frameshifts, fusions and copy number variants that have been clinically observed or curated from scientific literature to help users better understand and define precise function and actionability.
Available as a web-based application, this expert-curated resource contains content from over 547,000 real-world clinical oncology cases combined with content from the QIAGEN Knowledge Base (QKB), providing gene-level, alteration-level, and disease-level information.

When a variant has been “clinically observed,” it means our professional clinical interpretation service has encountered this alteration in a real-world clinical case.
For these variants, our team has assessed the clinical and biological relevance and calculated the gene and variant prevalence across observed tumor types.
Conversely, content from the QKB is proactively curated from scientific literature; therefore, not all variants have yet been directly clinically observed by our professional clinical interpretation services.
HSMD leverages expert-curated content from the QIAGEN Knowledge Base, which contains the latest evidence from peer-reviewed papers, clinical and functional studies, on- and off-label drugs, and professional guidelines, such as NCCN, AMP/ASCO/CAP and the World Health Organization (WHO), and commonly used databases, including gnomAD and ClinVar.
With this content, you gain insights into:


COSMIC, the Catalogue of Somatic Mutations in Cancer, is the world’s largest expert-curated somatic mutation database. HSMD complements COSMIC with data from real-world oncology cases to better understand gene and variant prevalence. HSMD also provides deeper variant annotations. Therefore, COSMIC is used to identify biomarkers and annotate variants for genomic reports, whereas HSMD is used to validate biomarkers and better assess their biological and clinical relevance.

Explore HSMD's features, content and applications with complimentary 5-day trial.